2022 year in review
2022 was a big year for JBrowse 2!
We started the year with v1.5.5 and ended with v2.3.2. Some big things happened in between!
Let's check some new features out.
New pre-prints
In 2022, we published preprints for our main JBrowse 2 paper https://www.biorxiv.org/content/10.1101/2022.07.28.501447v1 and JBrowse Jupyter https://www.biorxiv.org/content/10.1101/2022.05.11.491552v1.
URL query parameter API
We added the ability to control JBrowse 2 using URL parameters in v1.6.4
This allows users to write URLs such as
http://yoursite/jbrowse/?loc=chr1:1,000,000-2,000,000&assembly=hg19&tracks=genes
This improves the ability to link to direct locations of interest from gene pages for example. See https://jbrowse.org/jb2/docs/urlparams/ for more info.
Navigating synteny using the linear genome view
In v2.2.1, we added the ability to view "SyntenyTracks" as a regular linear genome view track. These tracks let you right click a feature and select "Open synteny view at this position" and it will show you e.g. the matching region on another assembly.
Base pair exact rendering of synteny
A long standing goal has been to render "base pair exact synteny" when the information is available e.g. as CIGAR strings.
This allows us to see that a "deletion relative to one genome" is an "insertion relative to the other".

The 'curvy' synteny line rendering mode was also added.
Floating labels adding to gene glyphs
In earlier versions, the label for a feature would not be visible if you were zoomed e.g. into the middle of the gene. In v1.7.0 we adding "floating labels" so that the feature label was always visible as you scroll.

Improved SVG export using canvas2svg
We switched to a battle tested canvas2svg library in v1.7.9 for exporting SVGs which improved exporting our 'sashimi-style' arcs on RNA-seq.

Feature detail panel customization
In v1.7.10, we added the ability to customize the feature details panel with custom links. This is very useful to add links to external resources or gene pages for different features.

See https://jbrowse.org/jb2/docs/config_guides/customizing_feature_details/ for more info
Upgrading to webpack 5 and reducing bundle sizes, and MUI v5 upgrade
In v1.7.0, we upgraded to webpack 5 which improved our bundle sizes, which makes the app load faster and feel more lightweight. In fact, over the lifetime of jbrowse 2, we have decreased our bundle size, and the initial load of v2.3.2 is actually 2.5x smaller than v1.0.0!

We also upgraded to MUI v5, which kept us on track with the latest and greatest from MUI, who continues to improve their UI widgets like the data grid.
Improved performance of the alignments tracks
We worked to improve alignments track performance throughout the year on a number of fronts. We greatly improved performance of the alignments track when web workers were used by avoiding serialization of all BAM/CRAM data (see https://jbrowse.org/jb2/blog/2022/04/26/v1.7.5-release/) and also did low level optimizations in our @gmod/bam and @gmod/cram libraries. There is still more to do, but we hope this improves user experience looking at large data files!

Figure showing BAM/CRAM performance on long and short reads of different coverages in JBrowse 2 versions over 2022. Figure generated by https://github.com/cmdcolin/jb2profile
(note: we also added web worker support to our embedded linear-genome-view, developers can manually opt-in to this with web worker RPC https://jbrowse.org/storybook/lgv/main/?path=/story/using-webworker-rpc--page)
More plugins for the plugin store!
We have continued see new plugins in the plugin ecosystem, including:
-
MultiLevelLinearView - a plugin that synchronizes multiple linear genome views at multiple zoom levels, by @carolinebridge-oicr
-
Reactome - showing pathways using Reactome's pathway viewer, by @carolinebridge-oicr
-
RefGet API for fetching sequence info from external contributor Emilio Righi, who has also contributed in making Vue adapters for our React code
See more here, as well as screenshots below: https://jbrowse.org/jb2/plugin_store/

Video of the MultiLevelLinearView
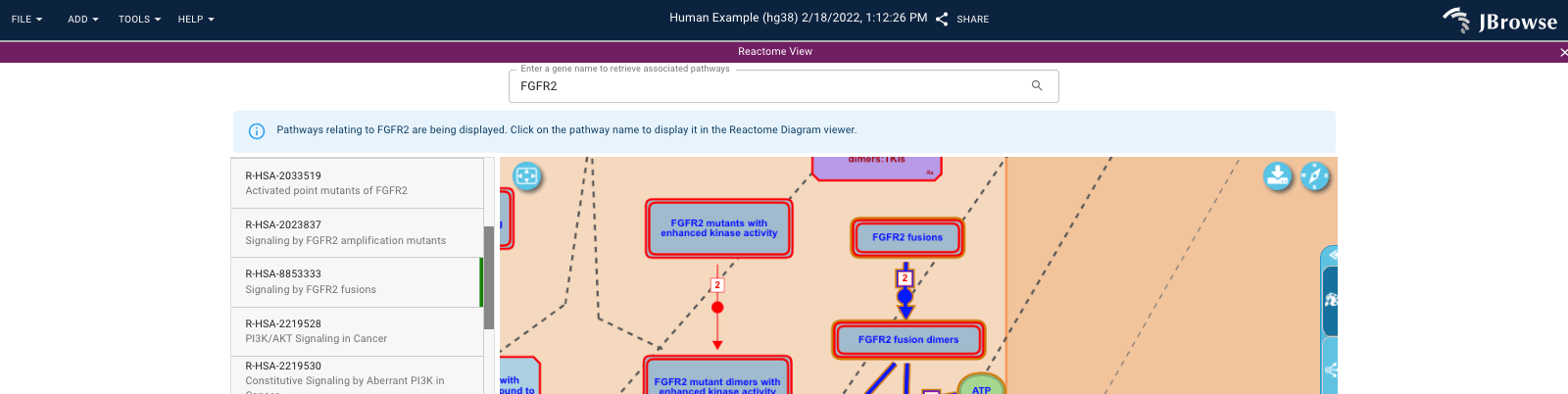
Screenshot of the Reactome plugin, showing the pathway viewer
Multi-wiggle tracks
We added the ability to have "multi-wiggle" tracks in v2.1.0, which let's you see multiple quantitative stopTokens on the same Y-scalebar easily.

"Overlaid" multi-wiggle track
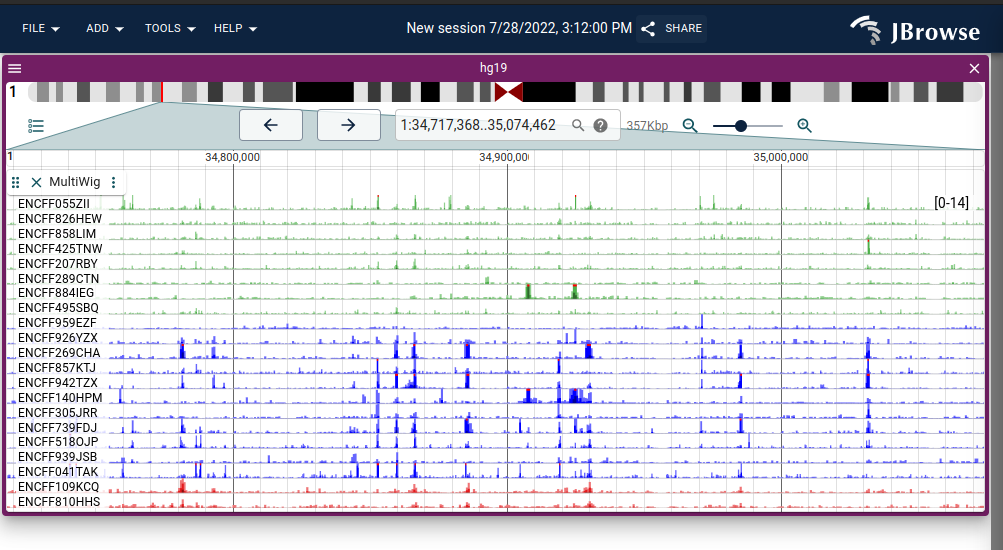
"Multi-row" multi-wiggle track
Arc and "read cloud" rendering of paired-end and split long reads
In v2.3.1, we introduced the ability to render the connection between paired-end and split-long reads using arcs! This style of view is similar to what we offered in the "breakpoint split view" but is more easily accessible being simply a linear genome view track rather than a custom view type!
We also created the "read cloud view" which stratifies the reads according to "insert size". Each of these features is similar to ones available in JBrowse 1, but has the added ability in JBrowse 2 to be able to connect across discontinuous regions (xref https://jbrowse.org/docs/paired_reads.html for JBrowse 1).
The features will color themselves according to read orientation (e.g. blue, teal, and green indicate various types of inversions or duplications) as well as insert size (pink is dynamically added if the insert size is statistically too small, which read is added if the insert size is statistically too large, each using +/- 3sd as a measure).

Looking forward
In 2023, we will start to see beta versions of the new Apollo rewrite with JBrowse 2 integration. A new single cell data viewer plugin is also in the works. Adding systems for multi-way synteny, exporting data, and faceted track selectors are also underway. Also, feel free to let us know what features you want to see!
Many of our team members will be at the Plant and Animal Genome conference 2023, so we look forward to seeing you there!
- Colin Diesh (@cmdcolin)
- Garrett Stevens (@garrettjstevens)
- Scott Cain (@scottcain)
- Caroline Bridge (@carolinebridge-oicr)
- Robert Buels (@rbuels)
- Teresa De Jesus Martinez (@teresam856)
