Introduction
Welcome!
This tutorial will show you how to get a JBrowse 2 linear genome view embedded in a website. It is similar to a tutorial given a BCC2020, but has been updated to reflect the final, released version of the interface.
What is JBrowse 2
JBrowse 2 is a pluggable open-source platform for visualizing and integrating biological data. At its core, it is a genome browser, but it has also been built as an extensible platform to enable visualization of all kinds of biological data.
For those of you who have used the original JBrowse, you'll recognize many of the ideas and use cases JBrowse 2 was built around. JBrowse 2 is a complete rewrite of the original JBrowse, however, and so there are some differences. We hope that you will see that these differences, along with the modern web technologies that power JBrowse 2, enable JBrowse 2 to be an even more flexible and useful tool.
What does it mean to embed JBrowse 2?
JBrowse 2 is built around the idea of componentization, meaning that the different pieces that are developed can be re-used to create different final products. Our full web app, called JBrowse Web, can show not only traditional linear genome browser views, but can also show Circos-style circular plots, dotplots, comparative views, and more. It can also display multiple different views simultaneously.
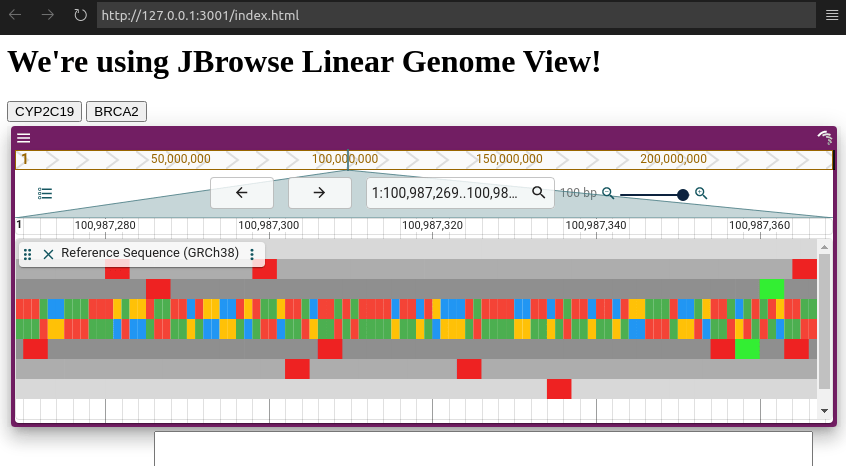
Today we'll be using JBrowse Linear Genome View, which has a subset of features of the full app. It can display a single linear view, which is the most common mode of genome visualization. Where JBrowse Web is a full-page web app, though, JBrowse Linear Genome View is designed to work within an existing web page. You can easily add one or several of these views to a page without the use of iframes, and control and react to each instance individually.
Who this tutorial is for
This tutorial is a good fit if you have an existing web page and you'd like to have JBrowse running inside that page, although in the tutorial we'll create a page from scratch so you don't need an existing page to do this tutorial.
If your web page is built with React, you can install our JBrowse Linear Genome View React Component from NPM instead of adding it like is done in this guide. A lot of the content of this tutorial would still be relevant to setting up and configuring the view, though.
Also, another option for your web site is to add the full JBrowse Web app as a new page in your site. This might be an option if the JBrowse Linear Genome View is missing features you need.
What you need
We recommend having Node.js installed so that you can have an easy way to run a server as well as be able to install the JBrowse CLI. Node.js has various installers here, or if you are on Mac or Linux and plan on doing more extensive JavaScript development, we recommend using NVM.
You can do most of this tutorial with a simple text editor and some way to serve
files (just opening the HTML files we create in a browser won't work, you'll
need a server). If you have Node.js installed, you can run a simple server by
opening your terminal in the directory you want to serve and running npx serve
(or you can install it globally with npm install -g serve and then run
serve).
We'll also be using the JBrowse CLI, although you can technically complete the
tutorial without it. You can install it after installing Node.js by running
npm install -g @jbrowse/cli. Check that it installed properly by running
jbrowse --help.