Beginnings of a simple site
Create a simple web page
Let's get started! The first thing we're going to do is create a simple web page into which we can embed JBrowse Linear Genome View. First, create a folder to the files in. Inside that folder, create a new file called "index.html" and open it in your preferred text editor/IDE. Paste the following into the file and save it:
<html>
<body>
<h1>We're using JBrowse Linear Genome View!</h1>
</body>
</html>
Start the server
Open your terminal and navigate to the folder where you saved your "index.html".
From there, run the command npx serve. It should print out a message that
looks something like this:
Serving!
Local: http://localhost:5000
Copied local address to clipboard!
Now open your web browser and navigate to the url (e.g. http://localhost:5000) You should see a web page that says "We're using JBrowse Linear Genome View!". If so, congrats, you're on your way to adding JBrowse Linear Genome View to a web site!
Add JBrowse
To add JBrowse Linear Genome View, you need to add the source to your page and
then render the component. Since JBrowse Linear Genome View uses React for
rendering, you'll also need to load the React source, too. We'll start by adding
a head to our page and use it to load the sources and a div that will hold
JBrowse. Update your "index.html" so matches what's below, then save it, switch
over to your browser, and refresh the page.
<html>
<head>
<script
src="//unpkg.com/react@16/umd/react.development.js"
crossorigin
></script>
<script
src="//unpkg.com/react-dom@16/umd/react-dom.development.js"
crossorigin
></script>
<script
src="//unpkg.com/@jbrowse/react-linear-genome-view/dist/react-linear-genome-view.umd.development.js"
crossorigin
></script>
</head>
<body>
<h1>We're using JBrowse Linear Genome View!</h1>
<div id="jbrowse_linear_genome_view"></div>
</body>
</html>
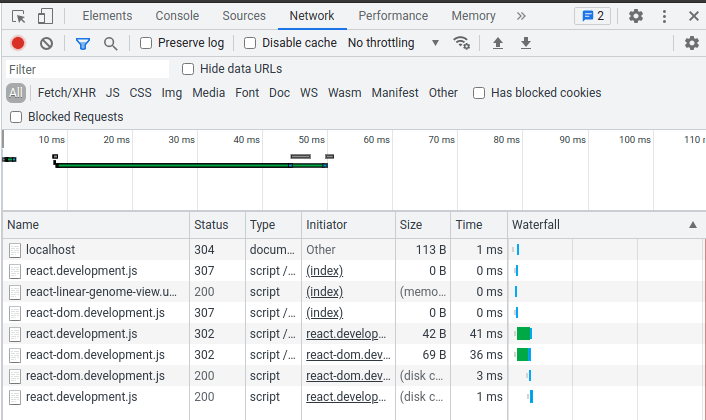
Now we can check if the script loaded properly. In your browser, open the developer tools (You can use F12 or Ctrl+Shift+I or right-click the page and select "Inspect") and go to the "Network" tab. If you see something like the below with a status of 200 for "react-linear-genome-view.umd.development.js," "react-dom.development.js," and "react.development.js," then you are good to go!
Now we need to actually create the view. The code to do so looks like this:
const { createViewState, JBrowseLinearGenomeView } =
JBrowseReactLinearGenomeView
const { createElement } = React
const { render } = ReactDOM
const state = new createViewState({
assembly: {
/* assembly */
},
tracks: [
/* tracks */
],
configuration: {
/* extra configuration */
},
plugins: [
/* runtime plugin definitions */
],
defaultSession: {
/* default session */
},
location: '', // location
onChange: () => {
/* onChange */
},
})
render(
createElement(JBrowseLinearGenomeView, { viewState: state }),
document.getElementById('jbrowse_linear_genome_view'),
)
First, though, we need to know what to put in all these options. Let's explore that next.