v2.3.1 Release
Hello all! This release adds some great new features!
-
New "arc" and "read cloud" alignments track modes which render long range connections between read pairs/split reads. These views have a big impact on being able to visualize structural variants.
-
We now automatically optimize the "prefix size" for our trix indexes created by
jbrowse text-indexwhich should help solve issues with slow text searching. -
We now refer to many pluggable elements by a "display name" instead of the coded class name so we have instead of "LinearGCContentDisplay" -> "GC content display".
New alignments track displays
We created a new display mode that changes the "pileup" of reads into "arcs" which connects both paired-end reads and split-long read alignments.

New track menu showing the ability to replace the "lower panel" with arc display or read cloud display

The arc view and read cloud view rendering across discontinuous displayedRegions

A zoomed in view of long-reads (top) and paired-end reads (bottom) at the breakpoint of a large deletion
Easily toggleable "compact" view of alignments
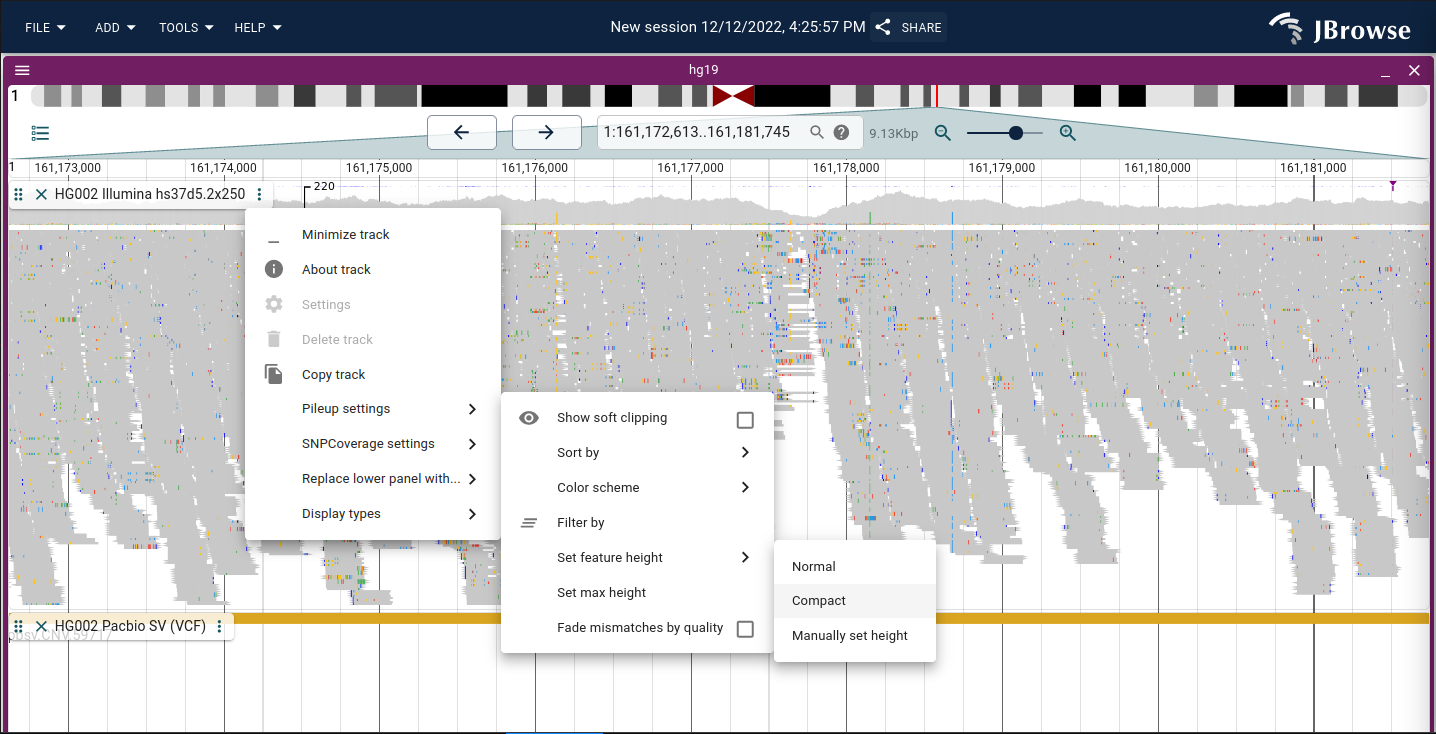
We now have an easily toggleable compact setting on alignments tracks (previously had to manually change feature height)
Optimized prefix size for text search indexing
Previously, if indexing long gene IDs with jbrowse text-index it would often
be slow because the "trix" format is generally optimized for short gene symbols.
We added the --prefixSize parameter in previous jbrowse versions to allow
optimizing for specific gene ID lengths, but this was sort of a magic number.
Now, jbrowse text-index will automatically calculate the --prefixSize if none
is provided, which tries to get an even ~64kb bin size. Let us know how it goes
for your data!
Re-install the CLI tools with e.g. npm install -g @jbrowse/cli to get the
latest version with this feature.
Moved "Track Hub Registry" plugin to plugin store
We removed the "Track Hub Registry" plugin, which was previously a "core plugin", to the plugin store. This will allow us to update the plugin over time and respond better to issues when they change their remote API.
Downloads
To install JBrowse 2 for the web, you can download the link above, or you can use the JBrowse CLI to automatically download the latest version. See the JBrowse web quick start for more details.
🚀 Enhancement
- Other
coretext-indexing
🐛 Bug Fix
- Other
- #3407 Remove trackhub registry plugin from core plugins, moved to plugin store (@cmdcolin)
- #3406 Fix loading connection tracks from connections that don't specify assemblyNames in config (@cmdcolin)
- #3390 Fix positioning within large alignments for query->target LGV synteny navigation (@cmdcolin)
- #3388 Fix search result that matches synonyms that matches multiple locations (@cmdcolin)
core
📝 Documentation
core- Other
🏠 Internal
__mocks__,core,text-indexingcore
Committers: 1
- Colin Diesh (@cmdcolin)